Philosophy - what is dtool?¶
What problem is dtool solving?¶
Managing data as a collection of individual files is hard. Analysing that data will require that certain sets of files are present, understanding it requires suitable metadata, and copying or moving it while keeping its integrity is difficult.
dtool solves this problem by packaging a collection of files and accompanying metadata into a self contained and unified whole: a dataset.
Having metadata separate from the data, for example in an Excel spread sheet with links to the data files, it becomes difficult to reorganise the data without fear of breaking links between the data and the metadata. By encapsulating both the data files and associated metadata in a dataset one is free to move the dataset around at will. The high level organisation of datasets can therefore evolve over time as data management processes change.
dtool also solves an issue of trust. By including file hashes as metadata it is possible to verify the integrity of a dataset after it has been moved to a new location or when coming back to a dataset after a period of time.
It is possible to discover and access both metadata and data files in a dataset. It is therefore easy to create scripts and pipelines to process the items, or a subset of items, in a dataset.
What is a “dtool dataset”?¶
Briefly, a dtool dataset consists of:
- The files added to the dataset, known as the dataset “items”
- Metadata used to describe the dataset as a whole
- Metadata describing the items in the dataset
The exact details of how this data and metadata is stored depends on the
“backend” (the type of storage used). In other words a dataset is stored
differently on local file system disk to how it is stored in Amazon S3 object
store. However, the dtool commands and the Python API for interacting with
datasets are the same for all backends.
What does a dtool dataset look like on local disk?¶
Below is the structure of a fictional dataset containing three items from an RNA sequencing experiment.
$ tree ~/my_dataset
/Users/olssont/my_dataset
├── README.yml
└── data
├── rna_seq_reads_1.fq.gz
├── rna_seq_reads_2.fq.gz
└── rna_seq_reads_3.fq.gz
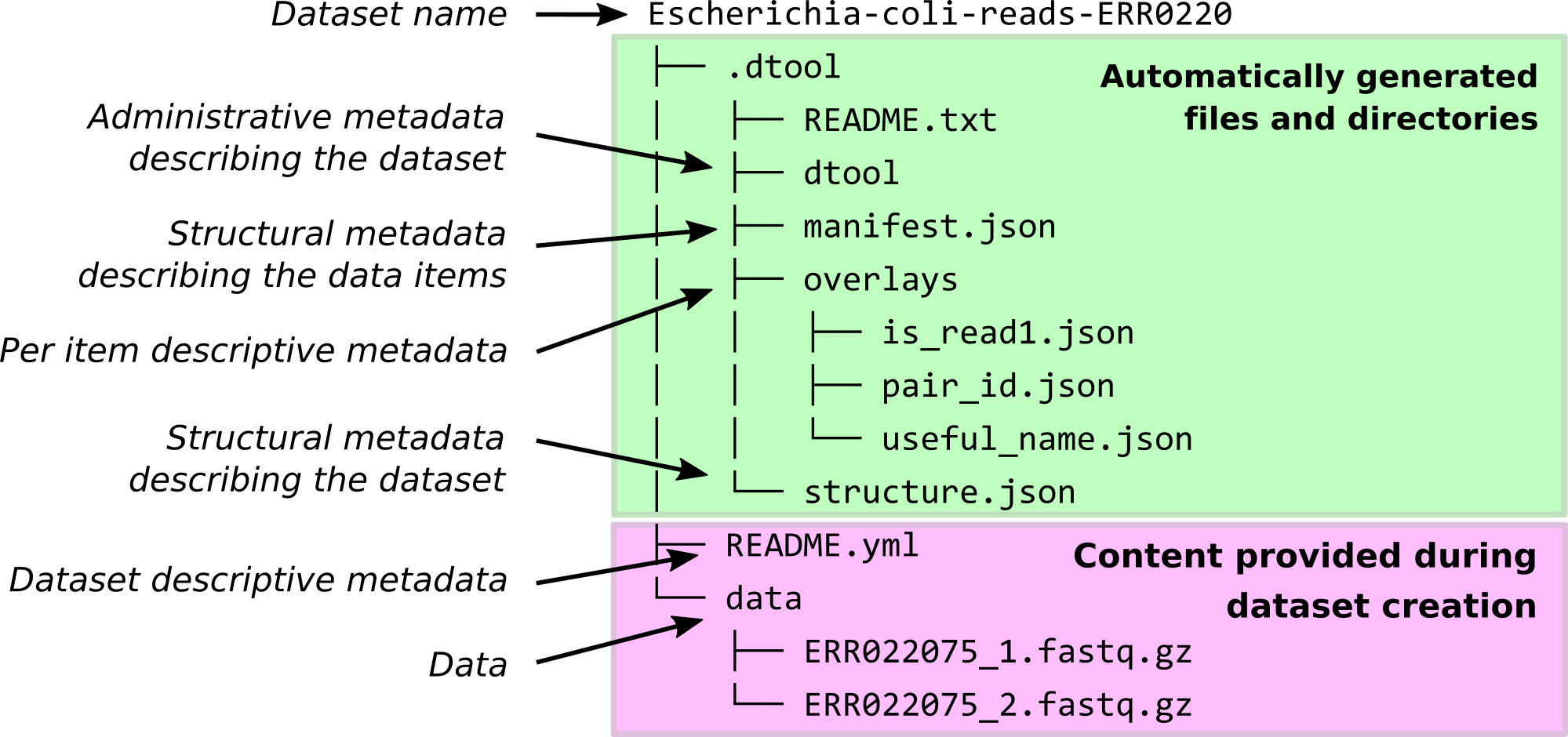
The README.yml file is where metadata used to describe the whole dataset is
stored. The items of the dataset are stored in the directory named data.
There is also hidden metadata, stored as plain text files, in a directory named
.dtool. This should not be edited directly by the user.
How does one create a dtool dataset?¶
This happens in stages:
- One creates a so called “proto dataset”
- One adds data and metadata to this proto dataset
- One converts the proto dataset into a dataset by “freezing” it
Once a proto dataset is “frozen” it is simply referred to as a dataset and it is no longer possible to modify the data in it. In other words it is not possible to add or remove items from a dataset or to alter any of the items in a dataset.
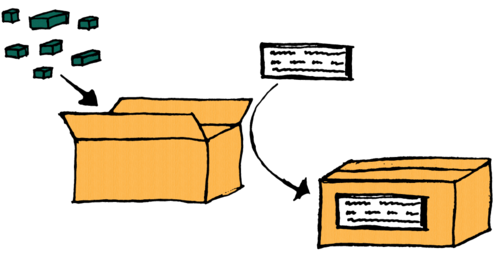
The process can be likened to creating an open box (the proto dataset), putting items (data) into it, sticking a label (metadata) on it, and closing the box (freezing the dataset).
Give me more details!¶
An in depth discussion of dtool can be found in the paper Lightweight data management with dtool.